englelab::get_template(score_script = TRUE)5 Score and Clean Data
Data Scoring
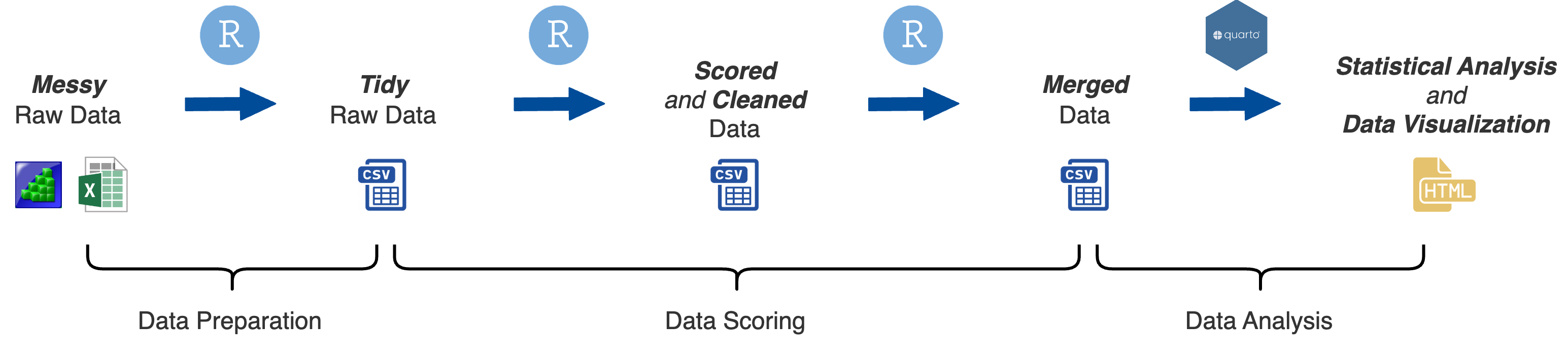
Before you can analyze the data you need to process the raw data into aggregate summary variables. This involves several steps:
Calculate task scores
Evaluate and remove problematic subjects and outliers
Calculate reliability
Overview of Template
You can download an R script template to score tidy raw data files:
Setup
# ---- Setup ------------------
# packages
library(here)
library(readr)
library(dplyr)
library(englelab) # for data cleaning functions
library(tidyr) # for pivot_wider. delete if not using
library(psych) # for cronbach's alpha. delete if not using
# directories
import_dir <- "data/raw"
output_dir <- "data/scored"
# file names
task <- "taskname"
import_file <- paste(task, "raw.csv", sep = "_")
output_file <- paste(task, "Scores.csv", sep = "_")
## data cleaning parameters
outlier_cutoff <- 3.5
# -----------------------------The Setup section is to:
Load any packages used in the script
Set the directories of where to import and output data files to
Set the file names of the data files to be imported and outputted
Set data cleaning parameters (e.g., outlier criterion)
Import
Given that 1) you created a tidy raw data file in .csv format, and 2) you specified import_dir and import_file in the setup section, you most likely do not need to change anything here.
Optionally, you might want to go ahead and filter out any rows that you absolutely do not want to include at any point in scoring the data (e.g., practice trials or certain subjects).
Score Data
The Score Data section is where most of the work needs to be done. You should be using dplyr and possibly tidyr to do most of the work here, though you may need other packages and functions. You can delete whatever is in there, that is just a placeholder as an example of the type of functions you might use.
# ---- Score Data -------------
data_scores <- data_import |>
summarise(.by = Subject)
# -----------------------------Clean Data
The next section of the script template is for cleaning the data by removing problematic subjects and/or removing outliers.
Reliability
There are two standard ways of calculating reliability: split-half and cronbach’s alpha. The script template provides some template code for calculating both of these.
Save Data
No need to change anything here. Isn’t that nice?
Tidy Data Example
Let’s use the tidy data created in the previous chapter (with two added participants)
data_import <- filter(data_import, TrialProc == "real")Trial-Level Cleaning
It can be a good idea to do some initial data cleaning at the trial level before scoring the data to make sure we are using only quality data.
The data cleaning that you need to do will of course depend on the nature of your data and study. Give some thought to the type of data cleaning steps you might want to take. Think about what would identify an observation as having low quality data (e.g., unrealistically short or long reaction times).
Reaction Times
For behavioral and cognitive tasks that include measures of reaction time, researchers will often exclude trials that have unrealistically fast reaction times or extremely long reaction times.
You will notice that in the example tidy raw data file, there are a few trials that contain unrealistically fast reaction times and extremely long reaction times.
A common criterion for unrealistically fast reaction times is anything less than 200 milliseconds. For extremely long reaction times, we can just use 5000 milliseconds (5 seconds) as the criterion.
There are two general strategies to do this:
use
filter()to get rid of the rows entirelyuse
mutate()andcase_when()to set the reaction time and accuracy values to missingNA
Using filter
# set criterion in setup section of script
rt_short_criterion <- 200
rt_long_criterion <- 5000
data_scores <- data_import |>
filter(RT >= rt_short_criterion, RT <= rt_long_criterion)Set to missing
Data Scoring
How you need to score your data depends on what you are measuring and what type of conditions you have. However, most approaches require calculating a summary statistic at some point. We will cover how to use summarise(.by = ) to calculate summary statistics.
With this example data let’s calculate a few different scores, aggregated by ID and Condition
Mean accuracy
Mean reaction time
Reaction time variability (e.g., standard deviation)
Number of trials aggregated over
# set criterion in setup section of script
rt_short_criterion <- 200
rt_long_criterion <- 5000
data_scores <- data_import |>
filter(RT >= rt_short_criterion, RT <= rt_long_criterion) |>
summarise(.by = c(ID, Condition),
Accuracy.mean = mean(Accuracy, na.rm = TRUE),
RT.mean = mean(RT, na.rm = TRUE),
RT.sd = sd(RT, na.rm = TRUE))Pivot
Now, depending on how this data is going to be analyzed (and even what statistical software you are using to analyze the data) you may want this data frame in either long (its current form) or wide format.
Between-Subjects ANOVA
Condition is not a between-subject variable in this sample data but if you have a between-subject variable or design, you will want to keep the column(s) for that variable(s) in long format.
Within-Subjects ANOVA
Condition is a within-subject variable in this sample data. Whether you want this variable in long or wide format depends on whether you will use R or JASP to analyze the data.
For R, you can keep it in long format
For JASP, you will need to restructure it to wide format
# set criterion in setup section of script
rt_short_criterion <- 200
rt_long_criterion <- 5000
data_scores <- data_import |>
filter(RT >= rt_short_criterion, RT <= rt_long_criterion) |>
summarise(.by = c(ID, Condition),
Accuracy.mean = mean(Accuracy, na.rm = TRUE),
RT.mean = mean(RT, na.rm = TRUE),
RT.sd = sd(RT, na.rm = TRUE)) |>
pivot_wider(id_cols = ID,
names_from = Condition,
values_from = c(Accuracy.mean, RT.mean, RT.sd),
names_glue = "{Condition}_{.value}")Use the small arrow ▸ at the end of the column names to see more columns
Correlation and Regression
For correlation and regression you will want to restructure the data to wide format (same as the code above).
If you have multiple data files from different measures or tasks, that you eventually want to merge into a single data frame for analysis, it is a good idea to add the measure or task name to the column names. That way when you merge the data you know which column corresponds to which task.
# set criterion in setup section of script
rt_short_criterion <- 200
rt_long_criterion <- 5000
data_scores <- data_import |>
filter(RT >= rt_short_criterion, RT <= rt_long_criterion) |>
summarise(.by = c(ID, Condition),
Accuracy.mean = mean(Accuracy, na.rm = TRUE),
RT.mean = mean(RT, na.rm = TRUE),
RT.sd = sd(RT, na.rm = TRUE)) |>
pivot_wider(id_cols = ID,
names_from = Condition,
values_from = c(Accuracy.mean, RT.mean, RT.sd),
names_glue = "{task}_{Condition}_{.value}")Use the small arrow ▸ at the end of the column names to see more columns
Subject-Level Cleaning
The next section of the script template is for cleaning the data by removing problematic subjects and/or removing outliers.
# ---- Clean Data --------------------------------------------------------------
data_cleaned <- data_scores |>
remove_problematic(
filter = "", # add a statement that would go in dplyr::filter
log_file = here("data/logs", paste(task, "_problematic.csv", sep = ""))) |>
replace_outliers(
variables = c(),
cutoff = outlier_cutoff,
with = "NA",
log_file = here("data/logs", paste(task, "_outliers.csv", sep = ""))) |>
filter(!is.na())
# ------------------------------------------------------------------------------Remove Problematic Subjects
Depending on the task, problematic subjects can be detected in different ways. For this example data we will simply remove subjects that had less than chance performance on congruent trials (were just guessing or did not understand the task).
To do so, we will use a custom function created for this purpose, remove_problematic() from our englelab package. ?englelab::remove_problematic
The main argument is remove =. This argument takes a logical statement (e.g., remove = Accuracy.mean <= .5).
The other argument, that is more optional, is log_file =. This allows us to save a data file containing only the subjects that were removed. This is good if we later on want to report in publications how many subjects were removed.
# set criterion in setup section of script
acc_criterion <- .34
data_cleaned <- data_scores |>
remove_problematic(
remove = "Stroop_congruent_Accuracy.mean <= acc_criterion",
log_file = here("data/logs", paste(task, "_problematic.csv", sep = "")))Use the small arrow ▸ at the end of the column names to see more columns
Remove Outliers
Remove outliers based on their final task scores. A typical way we remove outliers is by setting their score to missing NA if it is 3.5 standard deviations above or below the mean.
To do so, we will use a custom function created for this purpose, replace_outliers from our englelab package.
There are several arguments that need to be defined. See ?englelab::replace_outliers for a description on this.
The fully piped |> code for this entire section looks like:
# set criterion in setup section of script
acc_criterion <- .34
outlier_criterion <- 3.5
data_cleaned <- data_scores |>
remove_problematic(
remove = "Stroop_congruent_Accuracy.mean <= acc_criterion",
log_file = here("data/logs", paste(task, "_problematic.csv", sep = ""))) |>
replace_outliers(
variables = "Stroop_incongruent_RT.mean",
cutoff = outlier_criterion,
with = "NA",
pass = 1,
id = "ID",
log_file = here("data/logs", paste(task, "_outliers.csv", sep = ""))) |>
filter(!is.na(Stroop_congruent_RT.mean))Problematic subjects removed: 1Outliers detected (pass = 1): 0Notice that filter(!is.na()) is specified after replace_outliers. This removes any of the outliers from the dataframe.
Use the small arrow ▸ at the end of the column names to see more columns
Calculate Reliability
There are two standard ways of calculating reliability: split-half and cronbach’s alpha.
It is best to calculate reliability only on the subjects that made it passed data cleaning.
reliability <- data_import |>
filter(ID %in% data_cleaned$ID) |>
mutate(.by = ID,
Trial = row_number())Use the small arrow ▸ at the end of the column names to see more columns
Split-half reliability
First trials need to be split between even and odd trials (or whatever split-half one is using).
Then scores can be re-calculated based on the even/odd splits. Everything that was done to score the data needs to be included, such as trial-level cleaning.
splithalf <- reliability |>
mutate(.by = ID,
Split = ifelse(Trial %% 2, "odd", "even")) |>
filter(RT >= rt_short_criterion, RT <= rt_long_criterion) |>
summarise(.by = c(ID, Condition, Split),
RT.mean = mean(RT, na.rm = TRUE)) |>
pivot_wider(id_cols = ID,
names_from = c(Condition, Split),
values_from = RT.mean)Use the small arrow ▸ at the end of the column names to see more columns
Finally, spearman-brown corrected split-half reliability can be calculated and added to the data frame
splithalf <- reliability |>
mutate(.by = ID,
Split = ifelse(Trial %% 2, "odd", "even")) |>
filter(RT >= rt_short_criterion, RT <= rt_long_criterion) |>
summarise(.by = c(ID, Condition, Split),
RT.mean = mean(RT, na.rm = TRUE)) |>
pivot_wider(id_cols = ID,
names_from = c(Condition, Split),
values_from = RT.mean) |>
summarise(r = cor(incongruent_even, incongruent_odd)) |>
mutate(r = (2 * r) / (1 + r))
data_cleaned$Stroop_incongruent_RT.mean_splithalf <- splithalf$rUse the small arrow ▸ at the end of the column names to see more columns
Cronbach’s alpha
Cronbach’s alpha is easier to calculate because we do not need to re-calculate the scores.
The first step is to get the data frame setup such that trial number is spread across columns
cronbachalpha <- reliability |>
filter(RT >= rt_short_criterion, RT <= rt_long_criterion) |>
select(ID, Trial, Accuracy) |>
pivot_wider(id_cols = ID,
names_from = Trial,
values_from = Accuracy)Use the small arrow ▸ at the end of the column names to see more columns
Then we can use psych::alpha() to calculate the average correlation between items and save it to the data frame
cronbachalpha <- reliability |>
select(ID, Trial, Accuracy) |>
pivot_wider(id_cols = ID,
names_from = Trial,
values_from = Accuracy) |>
select(-ID) |>
alpha() # from the psych packageSome items ( 1 ) were negatively correlated with the total scale and
probably should be reversed.
To do this, run the function again with the 'check.keys=TRUE' optiondata_cleaned$Stroop_incongruent_RT.mean_cronbachalpha <-
cronbachalpha$total$std.alphaUse the small arrow ▸ at the end of the column names to see more columns